This web page was produced as an assignment for Genetics 677, an undergraduate course at UW-Madison.
Uniprot Accession Number: P01106
Protein Sequence
>gi|71774083|ref|NP_002458.2| myc proto-oncogene protein [Homo sapiens]
MDFFRVVENQQPPATMPLNVSFTNRNYDLDYDSVQPYFYCDEEENFYQQQQQSELQPPAPSEDIWKKFEL
LPTPPLSPSRRSGLCSPSYVAVTPFSLRGDNDGGGGSFSTADQLEMVTELLGGDMVNQSFICDPDDETFI
KNIIIQDCMWSGFSAAAKLVSEKLASYQAARKDSGSPNPARGHSVCSTSSLYLQDLSAAASECIDPSVVF
PYPLNDSSSPKSCASQDSSAFSPSSDSLLSSTESSPQGSPEPLVLHEETPPTTSSDSEEEQEDEEEIDVV
SVEKRQAPGKRSESGSPSAGGHSKPPHSPLVLKRCHVSTHQHNYAAPPSTRKDYPAAKRVKLDSVRVLRQ
ISNNRKCTSPRSSDTEENVKRRTHNVLERQRRNELKRSFFALRDQIPELENNEKAPKVVILKKATAYILS
VQAEEQKLISEEDLLRKRREQLKHKLEQLRNSCA
The protein sequence was obtained from Entrez and is in the FASTA format.(6)
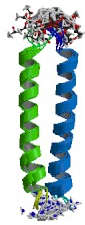
The above image is of the MYC-MAX heterodimer leucine zipper. The zipper is linked through a disulfide bond that is present at the N-termini of the protein.(5)
Protein Domain

Within the protein sequence of MYC, a helix-loop-helix (HLH-red in figure to left) domain was found to be present along with the amino terminal region (green) and a leucine zipper (yellow).(2) The HLH domain is contained between the 375 amino acid residue and the 427 residue. HLH domains consist mainly of basic residues which enables them to interact with negatively charged DNA. The HLH domain is found in eukaryotes and functions as a transcription factor in developmental pathways. The specific actions of these transcription factors is to cause a dimerization of inactive monomoers to a complex that acts in trans and enables development to continue in a highly regulated manner.(1) The presence of a leucine zipper domain is also indicative of MYC's ability to recognize and intreract with DNA. More specifically, MYC forms a heterodimer with the MAX protein. The formation of the MYC-MAX heterodimer is used to control cell growth and apoptosis and does so by interacting directly with genes that govern cell replication.(2)
MYC Inhibitor
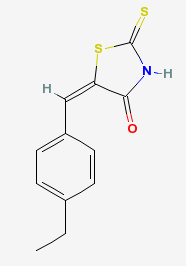
The image to the left is the structure of a chemical compound that inhibits the interaction between the MYC and MAX complex. The compound is a thiazolidinone that is capable of permeating the cell membrane and prevents the "transactivation of c-MYC target gene expression". C-myc inhibitor is capable of inhibiting MYC directed tumor cell growth. It has been shown to work both in vitro and in vivo and makes it a useful compound for studying the functions of myc and the resulting phenotypes. (3)(4) For another example of a compound that influences MYC, please visit the Scientific Article Review page.
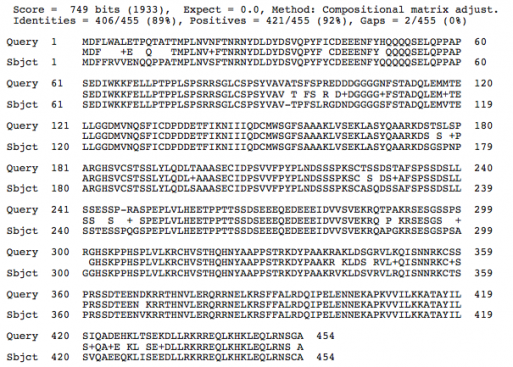
Comparison of Protein Structures
References:
1SMART
2Pfam
3NCBI-PubChem
4CalbioChem
5Protein Database
6Entrez
7Blast
8Homologene
Contact Info: Deeter Neumann, [email protected], May 14, 2009